Antibiotic-Resistant Superbugs Can Help Their Weaker Companions Survive And Thrive, Study Reveals
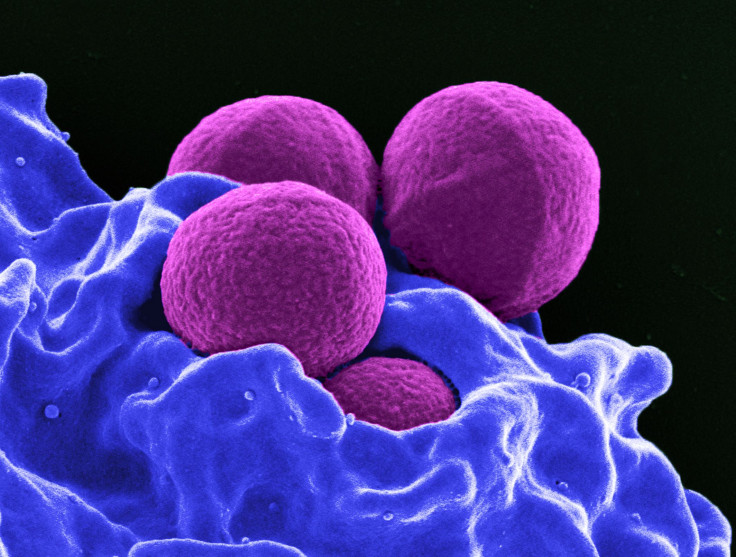
Scientists across the world have already raised alarm over the indiscriminate use of antibiotics and the associated rise in drug-resistant bacteria. Now, a new study, published Tuesday in the journal PLOS Biology, has unearthed a further complicating factor insofar as antibiotic resistance in bacteria is concerned.
The study revealed that bacteria that are susceptible to antibiotics can survive longer when in the company of their drug-resistant counterparts.
“We know that antibiotic usage results in selection for resistance,” lead author Robin Sorg, a microbiologist at the University of Groningen in the Netherlands, said in a statement. “However, we do not fully understand the processes, nor why antibiotic resistance can develop so fast. Single cell studies like ours help to fill in some of these details.”
For the purpose of their study, the researchers infected mice with a cocktail containing both — Staphylococci bacteria resistant to the antibiotic chloramphenicol and non-resistant Streptococci. They found that not only did the chloramphenicol-sensitive bacteria survive, they also began dividing — eventually outgrowing their resistant companions.
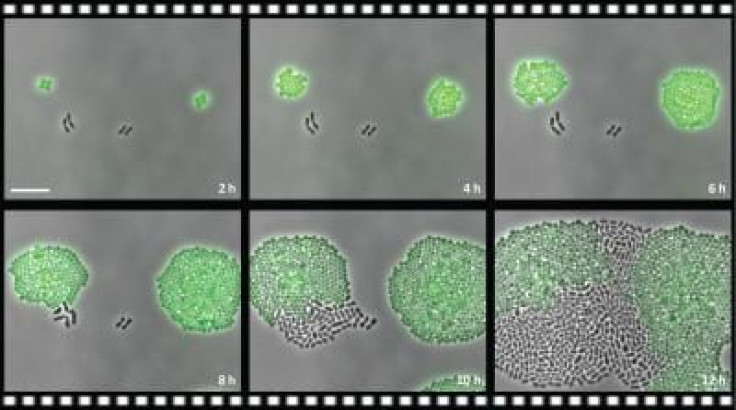
“We show that bacteria expressing the resistance factor chloramphenicol (Cm) acetyltransferase (CAT) can potently deactivate Cm in their immediate environment. The reduced Cm concentration then allows for the outgrowth of genetically susceptible bacteria in the same environment,” the researchers wrote in the study. “Our results highlight the importance of the microbial context during infectious disease as a potential complicating factor to antibiotic therapy.”
A similar phenomenon has been previously seen in bacteria resistant to penicillin, which secrete beta-lactamase enzymes to break down the antibiotic.
“But in our case, the antibiotic is deactivated inside the resistant cells,” Sorg said.
The worrying findings come at a time when indiscriminate and improper use of antibiotics has surged, and the rise of “superbugs” has become a serious global health concern. The World Health Organization, for instance, terms antimicrobial resistance “an increasingly serious threat to global public health that requires action across all government sectors and society.”
According to a recent British government-commissioned review, Antimicrobial Resistance (AMR), which already kills roughly 700,000 people every year, may, by 2050, lead to a staggering 10 million deaths annually.
“Even at current rates, it is fair to assume that over one million people will have died from AMR since I started this review in the summer of 2014,” economist Jim O'Neill — who was appointed by British Prime Minister David Cameron to head the review — said in the report.
© Copyright IBTimes 2024. All rights reserved.












